DoD funds CSCL to engineer killer cells!
See below for project details
Simulated actin waves
Department of Electrical and Computer Engineering
Johns Hopkins University

See below for project details
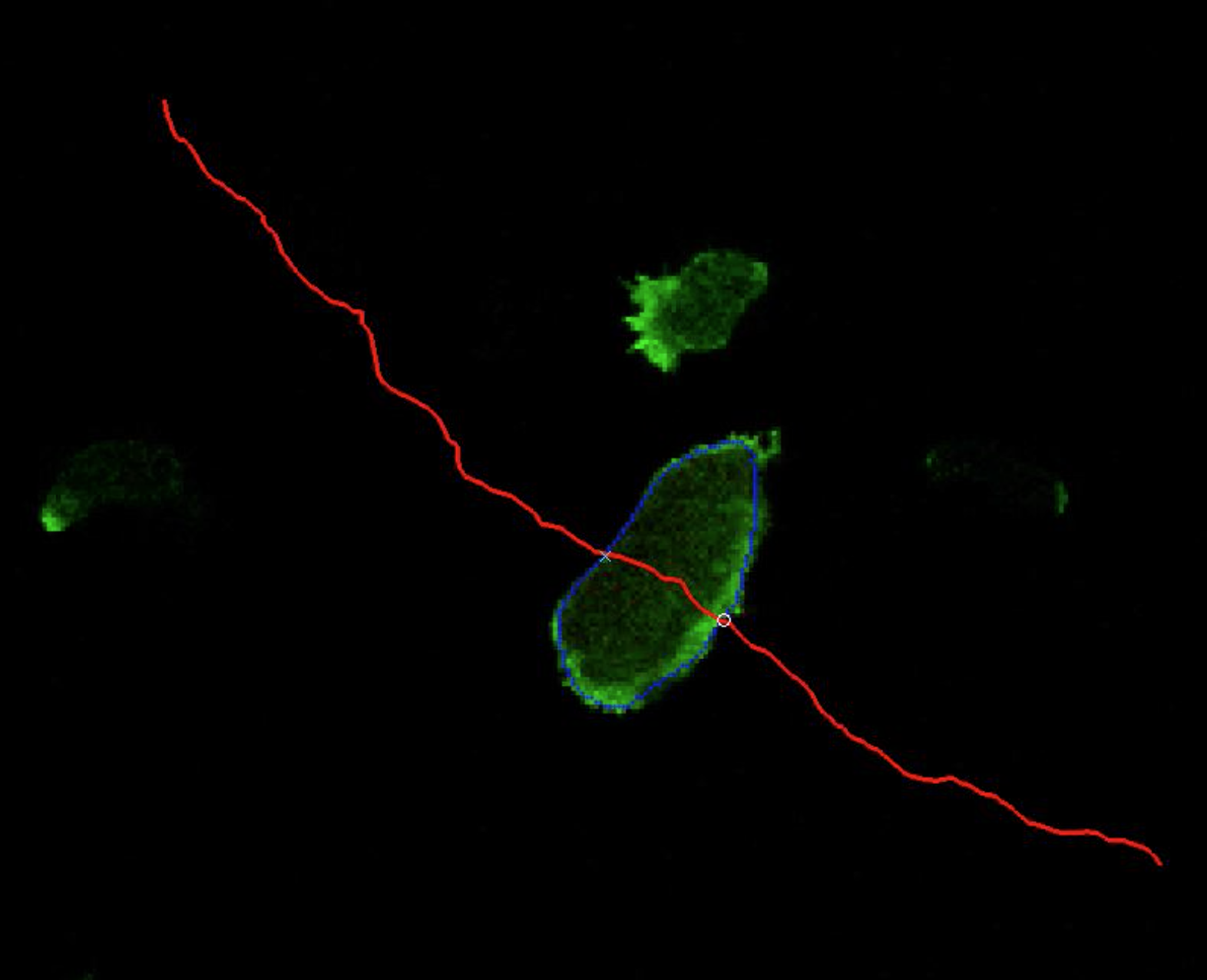
New publication, Nature Cell Biology 2017

New PhD student Kathleen DiNapoli joins the lab in March
Recognized for his contributions to control theory and systems biology

Ongoing project by the lab featured in the newspaper
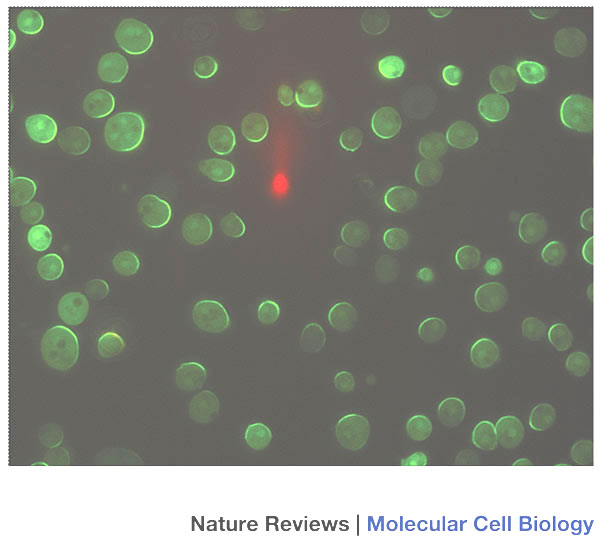
From the archive: The gradient sensing and motion model

Pablo's presentation at the Mathematical Biosciences Institute.
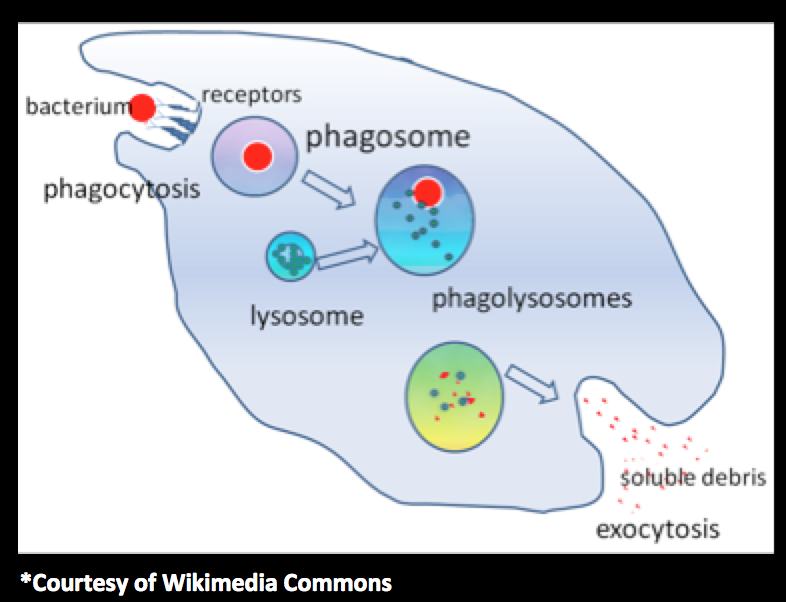
What we do
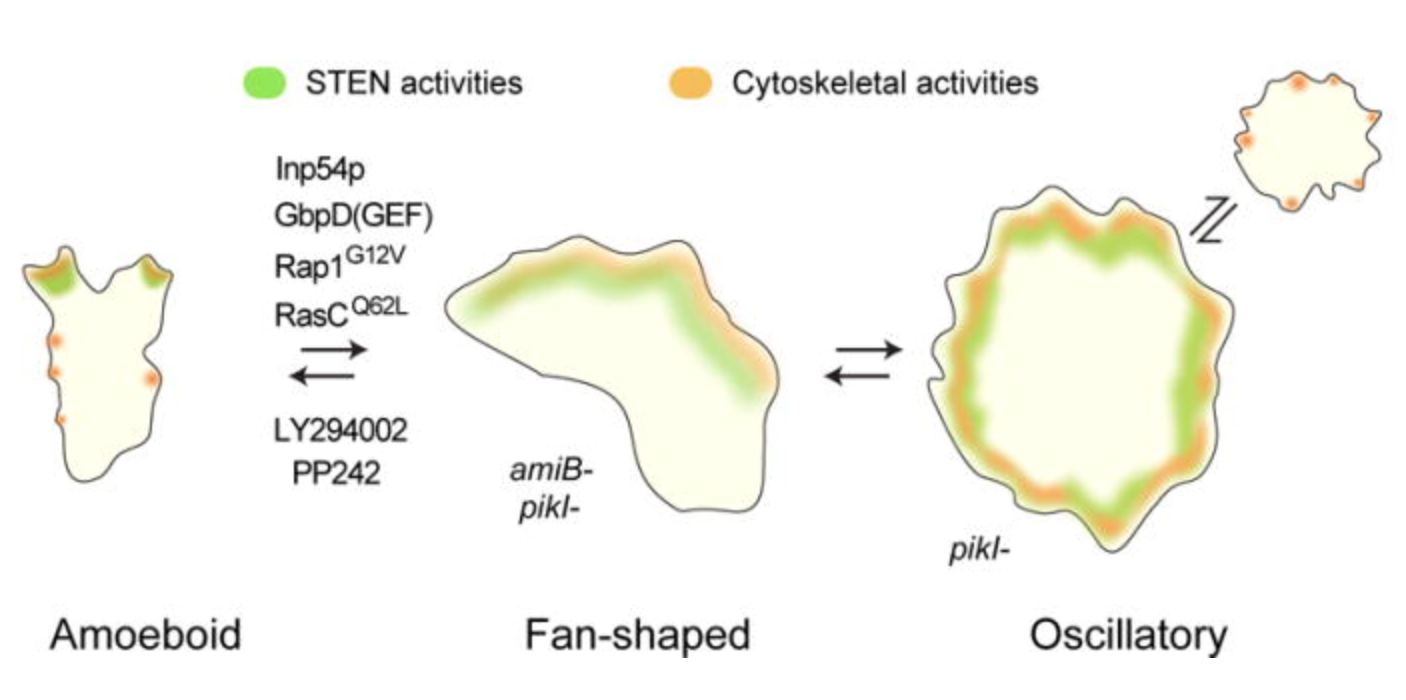
Creating models for faster-moving cells that can engulf pathogens with high efficiency. Collaborating with experimentalists in order to synthetically implement the models and engineer the Killer-Dicty.
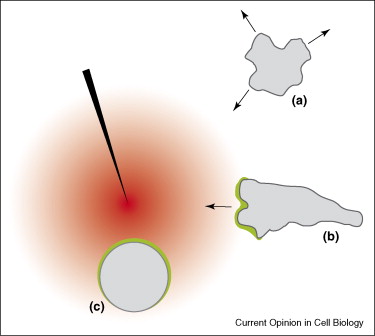
Controlling the dynamics of pseudopod formation during cell migration using theoretical models that analyze the phenomenon of wave propagation observed through actin waves at the cell cortex.
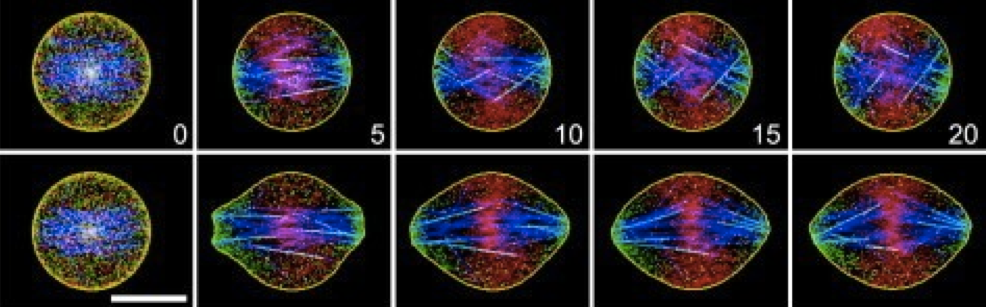
Modeling the dynamics of myosin and other contractile machinery during cell division. Establishing control mechanisms through the proteins responsible for contraction which can be used to create faster cells.
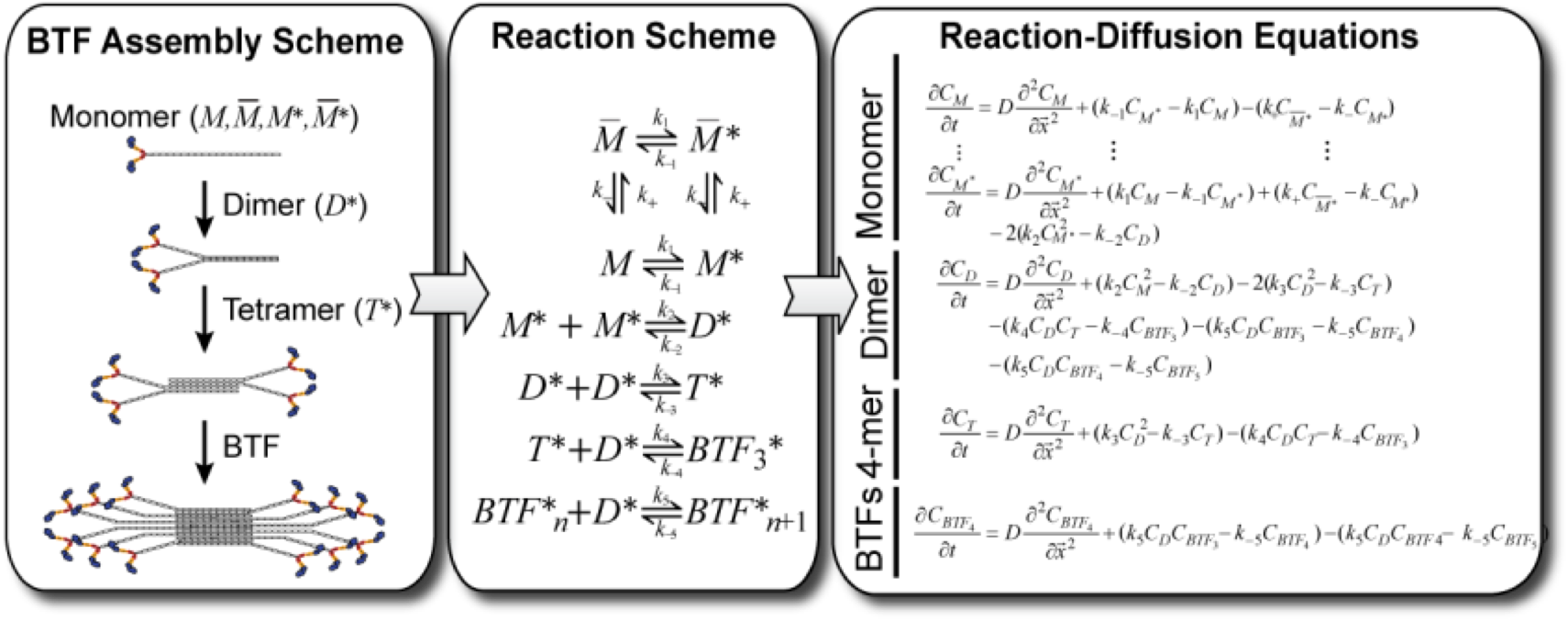
Deconstructing the pathways involved in cellular lipid signaling, such as the G-protein cascade and the PI3K-Akt pathway. Using detailed stochastic descriptions to match observed experiments in order create a picture of the complex signalling network.
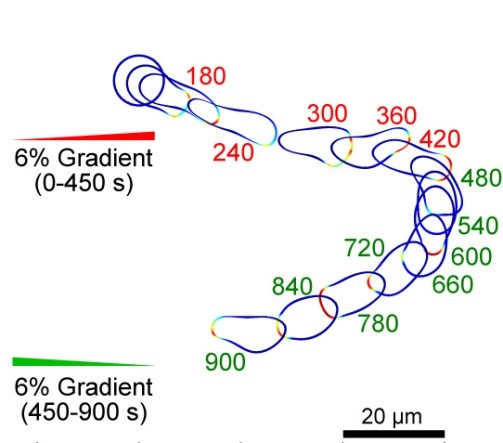
Using the level set framework to model cell shape changes during migration, cell-cell collision, cytokinesis, etc. Analyzing the implications of cell morphology changes for migration in different terrain.
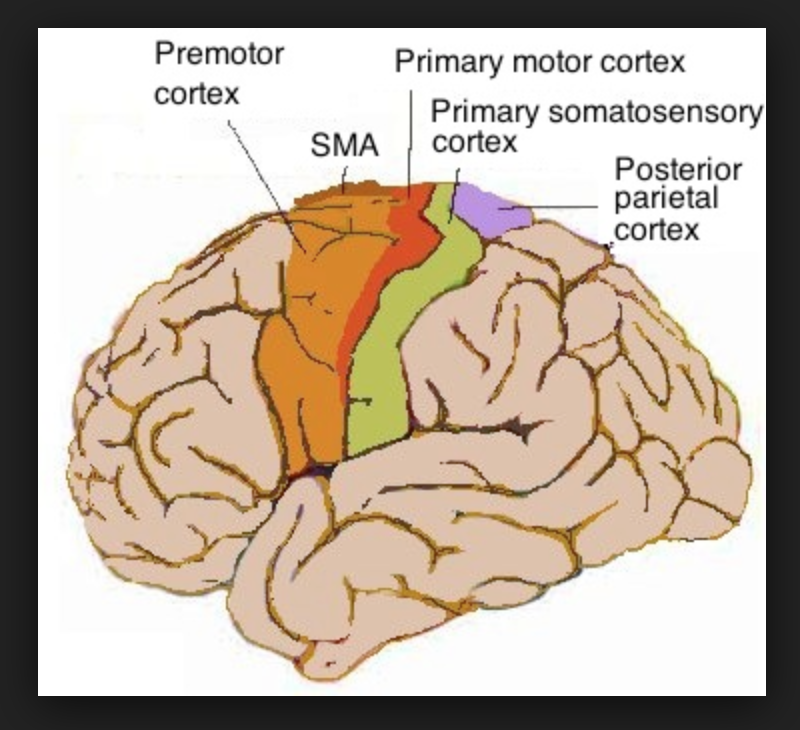
Modelling the wave propagation characteristics seen in the brain, in order to be able to predict behavioral changes in primates. Creating a layered architecture for the thalamocortical system in order to study connectivity properties.